Quickly Find Structural Elements by Name in SAMSON
Why Labels Disappear When You Zoom Out (And How to Control It in SAMSON)
How to Select Atoms with High Partial Charges in SAMSON Using NSL
When analyzing molecular systems, identifying atoms with high partial charges is often essential for understanding reactivity, electrostatic interactions, or molecular dynamics behavior. For example, atoms with partial charges above a certain threshold can be involved in crucial non-covalent interactions such…
Speed Up Nanostructure Design with Pattern Editors in SAMSON
Why Box Shape Matters: Choosing the Right Unit Cell in Molecular Simulations
Easily Select ARAP Active Atoms with Node Specification Language in SAMSON
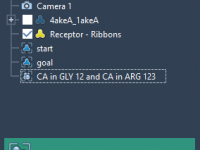
When modeling protein conformational transitions, choosing the right atoms to guide your simulations can make a big difference. In SAMSON’s Protein Path Finder app, using the ARAP (As-Rigid-As-Possible) method requires identifying specific active atoms—those that drive structural changes and influence…