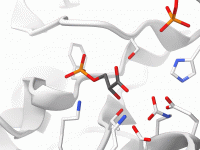
Molecular modeling often requires combining clarity with aesthetic quality. When working with complex biomolecular systems, it can be time-consuming to apply multiple visual layers manually — ribbons for secondary structures, sticks for ligands, van der Waals spheres for ions, color…