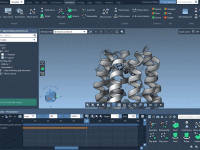
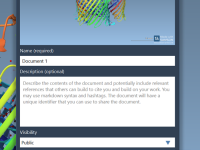
Tired of Emailing Molecular Data Back and Forth? Here’s a Better Way.
How to Set Up a Receptor Model for Docking in SAMSON—in 5 Minutes
How to Minimize Just Part of a Molecule in SAMSON Without Affecting the Rest
Accidentally deleted a molecule? Here’s how to recover it in SAMSON.
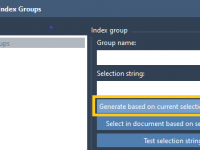
Quickly find non-hydrogen atoms with NSL
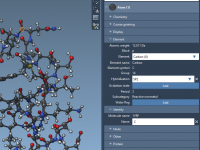
Quickly Modify Molecular Properties Using the SAMSON Inspector
Quick Presence, Quick Exit: Flash Animations for Molecular Storytelling
When you’re designing molecular presentations, timing is everything. Whether you’re teaching, presenting to colleagues, or preparing a video abstract for a publication, manually toggling the visibility of molecular nodes can be time-consuming—and often results in jarring, unnatural transitions. This is…