Improving Molecular Presentations with Background Transitions in SAMSON
Crafting clear, visual molecular presentations often requires more than just beautiful molecules — context is important. If you’ve ever found yourself wishing your molecular animations could flow more smoothly between informational slides or contextual backgrounds, SAMSON’s Set background animation can…
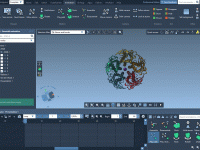
Getting Structures into Place: Using Assemble Animation in Molecular Presentations
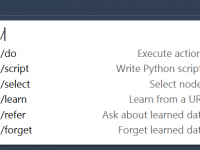
When ChatGPT Writes Your Python Scripts for Molecular Modeling
Avoid Simulation Bugs: Know Which Nodes Are Actually Visible
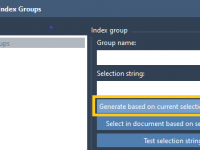
Stop Manually Clicking: A Cleaner Way to Select Files in SAMSON with NSL
No More Waiting: How to Run Molecular Simulations in the Cloud Without Overloading Your Computer
Why Some Nodes Disappear in Your Molecular Model (and How to Find Them Again)
Struggling to Manage Complex Molecular Projects? Document View Can Help
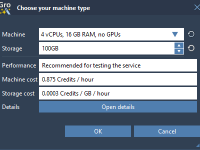
Running GROMACS Simulations in the Cloud without the Setup Stress
For molecular modelers, one recurring challenge is ensuring enough computational power to run molecular dynamics simulations efficiently. This becomes especially pressing when working with large systems or when comparison between multiple parameter combinations is needed. Local machines often fall short,…