Author: OneAngstrom
Why Molecular Modelers Should Care About SAMSON Apps
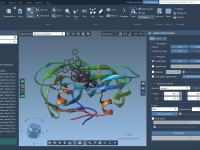
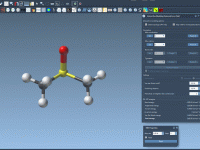
When Bonds Break — and Reform: Exploring Topology Changes with IM-UFF
Quickly Identifying the Right Render Presets in Complex Molecular Scenes
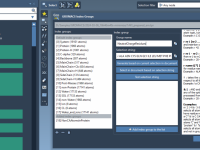
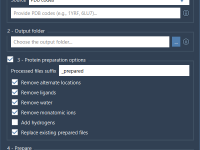
Custom Index Groups in GROMACS: A Simple Way to Target Specific Atoms and Residues
How to Quickly Find the Right Chains in Large Molecular Systems
Filtering Molecular Models by Atom Counts in SAMSON’s NSL
Custom Index Groups: Fine-Tune Your Simulation Setup in SAMSON
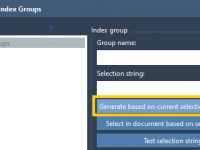
When preparing molecular simulations with GROMACS, setting up your system with precision can save you from troubleshooting downstream. One common pain point for molecular modelers, particularly when dealing with analysis or advanced simulation protocols like umbrella sampling and pulling experiments,…