Author: OneAngstrom
Shifting Perspectives: Horizontal Camera Movement in Molecular Presentations
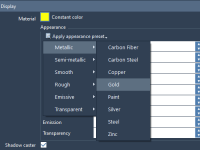
From Metal to Glass: How to Quickly Style Molecular Models with Materials in SAMSON
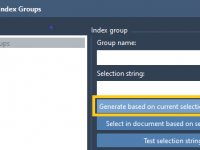
Easier GROMACS Selections with Custom Index Groups in SAMSON
Going from FASTA to 3D Structure in Minutes with AlphaFold-2 in SAMSON
Selecting Molecular Structures with Natural Language in SAMSON AI
Working with molecular structures often means managing complex selections—atoms, residues, chains, or interaction regions. For many researchers, this translates into repetitive interface navigation, complex queries using atom specifications, or scripting with a steep learning curve. But what if you could…