Author: OneAngstrom
Avoiding Common Pitfalls When Adding Ions to Coarse-Grained Systems in GROMACS Wizard
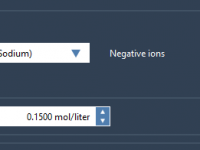
When preparing coarse-grained (CG) molecular systems for molecular dynamics (MD) simulations, one crucial step is the addition of ions. This step ensures charge neutrality and mimics physiological conditions. However, it’s easy to encounter unexpected issues—especially when working with CG models—if…
Installing Python Packages in SAMSON: A Quick and Flexible Workflow
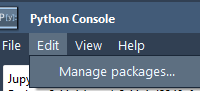
Whether you’re analyzing molecular dynamics, building neural network models for protein folding, or integrating your own Python toolkits, managing your Python packages efficiently is essential. Unfortunately, for many computational scientists, package installation can be frustrating. Switching between environments, worrying about…
Controlling Molecule Visibility in SAMSON with NSL Filters
Filtering Side Chains by Atom Composition in SAMSON
Better Molecular Scenes with Visual Presets in SAMSON
Molecular modeling often involves balancing scientific accuracy with visual clarity. Whether you’re preparing molecular graphics for a publication, presentation, or educational material, achieving visual consistency while highlighting important features can be time-consuming. This is where SAMSON’s Visual Presets come in.…
Tired of Atom Jumps in Molecular Animations? Keyframes Can Help
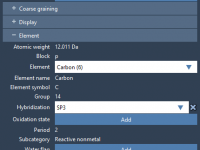
Editing Molecular Properties in SAMSON: A Closer Look at the Inspector
A Better Way to Pan Molecular Systems Horizontally
Quickly Find Molecular Structures with NSL in SAMSON’s Document View
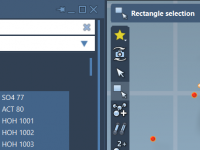
Molecular modelers often work with large, complex systems—think lengthy protein chains, multiple ligands, or layered structural hierarchies. Navigating and selecting specific components in such systems can easily become overwhelming. To address this challenge, SAMSON introduces the Node Specification Language (NSL),…