Author: OneAngstrom
When Designing Molecules in 2D is the Smartest First Step
Unlocking New Molecular Modeling Capabilities with SAMSON Apps
A Simpler Way to Track Molecular Simulations in the Cloud
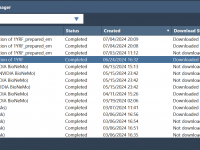
If you’re a molecular modeler juggling simulations across heavy-duty software like AlphaFold, GROMACS, or NVIDIA BioNeMo services, you’ve probably experienced the headaches of managing multiple jobs: locating results, checking progress, and syncing data across workstations. This can become tedious—especially if…
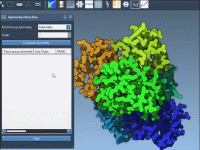
How to Select Terminal Residues in SAMSON with NSL
Why Your Ligand Path Sampling May Be Failing (and How to Fix It)
Easier Than You Think: Adding and Removing SAMSON Extensions

Many molecular modelers appreciate having specialized tools for specific simulations, from free energy estimation to advanced visualization techniques. But customizing your environment without wasting time or disrupting your scientific workflow can be a challenge. That’s where SAMSON Connect’s Extension Marketplace…