Author: OneAngstrom
Effortlessly Manage Cloud Jobs in Molecular Modeling
Enhancing Molecular Animations with the Hide Effect in SAMSON
Step-by-step: Building Lipid Layers Around Proteins in SAMSON
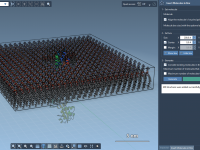
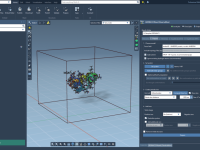
For molecular modelers, creating realistic lipid bilayers or membranes around proteins can be a time-consuming and intensive task. Whether you’re studying protein-membrane interactions or working on membrane protein systems, recreating lipid environments is crucial. The Molecular Box Builder extension in…