Mastering the ‘Reveal Atoms’ Animation for Molecular Modeling
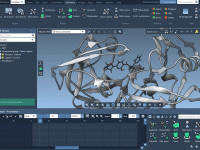
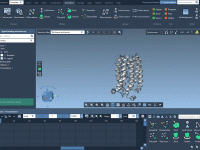
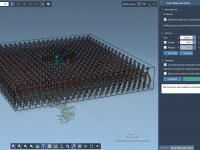
Molecular modeling often involves juggling intricate visualizations to present atomic-level insights effectively. One particular challenge revolves around highlighting specific atoms or molecular bonds progressively, beginning from zero visibility and transitioning towards a clear display. For molecular designers, this problem can…