Author: OneAngstrom
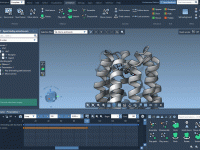
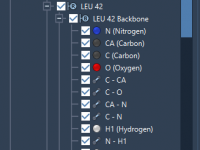
Mastering Light Attributes in Molecular Modeling with SAMSON
Effortlessly Follow Atoms During Molecular Simulations.
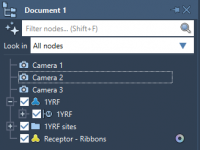
Mastering Molecular Visibility with the Flash Animation in SAMSON
For molecular modelers designing presentations or simulations, achieving visual clarity while managing the visibility of molecular components can be a challenge. Whether you’re illustrating a specific molecular process or emphasizing the dynamics of an interaction, focusing and removing elements at…
Streamlining Molecular Modeling with SAMSON Apps.
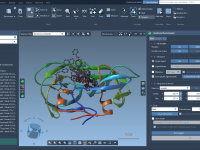
For molecular modelers, the ability to seamlessly integrate multiple tools and functionalities is often pivotal in solving complex problems. Whether you’re running simulations, visualizing molecular structures, or connecting with external tools and services, SAMSON’s app ecosystem provides a flexible and…