Author: OneAngstrom
Mastering Batch Computations with GROMACS Wizard
Performing molecular simulations often involves managing multiple conformations or preparing different molecular systems. This can be an overwhelming and tedious task when done manually. Fortunately, the GROMACS Wizard in SAMSON simplifies these processes through batch computations, allowing molecular modelers to…
Maintain a Static View with the Hold Camera Animation in SAMSON
Simplify Collaboration with Shared Documents in SAMSON

Collaborating effectively within scientific teams or across institutions can often feel challenging, especially when working on complex molecular modeling projects. Sharing results, models, and insights securely while ensuring compatibility is essential for success. SAMSON has an integrated solution for these…
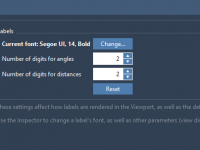
Enhance Your Molecular Models with Customized Labels
Best Practices for Uninstalling SAMSON
Master Molecular Visualization with Color Schemes in SAMSON!
Optimizing Molecular Simulations with Custom GROMACS Versions in SAMSON
Efficiently Colorize Residues in 3D with Sequence Views
Exploring Defects in Diamond Structures with SAMSON.
When modeling materials, understanding how defects influence a crystal’s properties is essential. Defects can significantly impact characteristics like strength, conductivity, and transparency. Luckily, SAMSON’s Crystal Creator offers a streamlined way to explore defects in crystals like diamond, making it easier…