Exploring Defects in Diamond Structures with SAMSON.
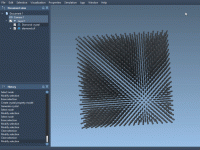
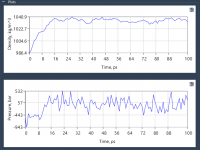
When modeling materials, understanding how defects influence a crystal’s properties is essential. Defects can significantly impact characteristics like strength, conductivity, and transparency. Luckily, SAMSON’s Crystal Creator offers a streamlined way to explore defects in crystals like diamond, making it easier…