Author: OneAngstrom
Simplifying Energy Minimization with SAMSON’s GROMACS Wizard.
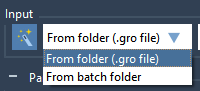
For molecular modelers, one major challenge is ensuring that molecular systems are geometrically stable before moving into simulations. This critical step—Energy Minimization—is essential to eliminate steric clashes and optimize the geometry of the system for further molecular dynamics simulations. With…
Mastering Simulation Parameters: A Key to Reliable Molecular Dynamics
Simplifying Molecular Conformation Analysis with NSL’s Attributes
Effortless Molecular Visualization with Color Schemes in SAMSON
Simplify Molecular Path Queries with SAMSON’s Path Attribute Space.
For molecular modelers dealing with conformations, querying specific paths efficiently can significantly speed up workflows and improve accuracy. SAMSON’s Path Attribute Space provides a streamlined way to define and manipulate path-related attributes, making molecular analysis more intuitive and precise. The…