Author: OneAngstrom
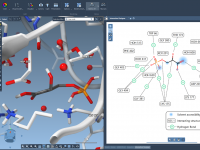
Seamlessly Sync 2D Interaction Diagrams with a 3D Viewport in SAMSON
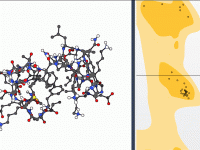
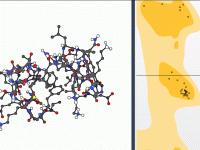
Master Dihedral Angles with the Interactive Ramachandran Plot Tool
Effortlessly Enhance Molecular Visualizations with Visual Presets in SAMSON
Creating clear and aesthetically pleasing molecular visualizations can often be a challenge for molecular modelers. Whether you are analyzing molecular structures or preparing visuals for a presentation or publication, the demand for high-quality imagery is constant. Thankfully, SAMSON’s powerful Visual…
Mastering Reverse Path Animations in Molecular Modeling
How to Configure Ligand and Protein States for Pathway Exploration
Understanding Light Attributes in Molecular Models.
From SMILES to 3D: Generating Molecular Structures with SAMSON’s SMILES Manager
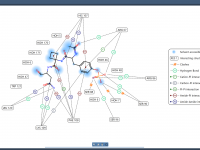
Streamlining Molecular Interactions: Creating 2D Interaction Diagrams with Ease
Streamlining Molecular Disassembly: A Guide to the Disassemble Animation in SAMSON
If you're a molecular modeler, you've likely encountered challenges when trying to visualize complex molecular systems. Whether it's understanding spatial arrangements or presenting your findings clearly, generating animations that simplify structural representations can be crucial in making your work accessible…