Author: OneAngstrom
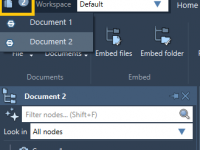
Understanding Node Types in SAMSON: A Practical Guide for Molecular Modelers
What You Need to Run SAMSON Smoothly
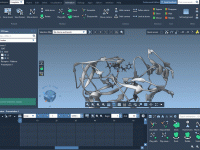
Enhancing Molecular Presentations with the Undock Animation
Ensuring Compatibility for Seamless Molecular Modeling
Effortlessly Record Atomic Trajectories in SAMSON
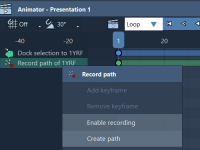
For molecular modelers, keeping track of atomic trajectories during presentations or simulations is often a crucial step. Whether you’re analyzing molecular movements, creating visualizations, or preparing a scientific presentation, precise trajectory tracking allows you to extract valuable insights. This is…