Making Molecular Systems Visually Intuitive with Visual Models in SAMSON

For molecular modelers, translating complex nanosystems into meaningful graphic representations is a recurring challenge. This is where SAMSON’s visual models come into play, offering comprehensive and customizable tools to bridge the gap between complex data and understandable visuals. Whether you’re…
Streamline Molecular Visualization with Light Attributes in SAMSON
For molecular modelers working with complex systems, managing visualization settings effectively can be a real challenge. The ability to control specific aspects of nodes—particularly light nodes—is crucial for understanding, presenting, and debugging molecular designs. This is where SAMSON’s light attributes…
Discover Transparent Transitions with the Pulse Animation in SAMSON
Enhancing Molecular Insights: Visualizing Protein-Ligand Docking Results in SAMSON
Streamline Molecular Modeling with Path-Specific Attributes in SAMSON
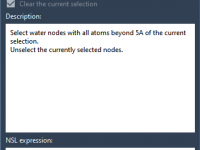
For molecular modelers, efficiently navigating and analyzing molecular conformations is key to unlocking insights. SAMSON’s Node Specification Language (NSL) provides advanced tools to address this challenge, particularly through the use of path attribute space. This attribute space, with the short…