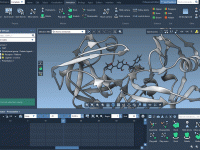
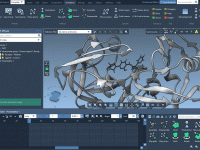
Mastering Folder Attributes in Molecular Modeling
Simplifying Molecular Dynamics: Applying Custom Parameters in SAMSON
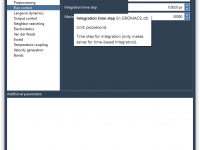
For molecular modelers, managing simulation parameters can sometimes feel overwhelming. Whether it’s energy minimization, equilibration, or production MD simulation, configuring the right settings for molecular dynamics often entails navigating through multiple files and tools. What if there was a way…
Simplifying Molecular Design with Key Property Model Attributes in SAMSON
Simplify Protein Transition Path Generation with ARAP Interpolation
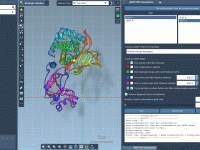
In the complex world of molecular modeling, creating realistic and biologically meaningful transition paths between different protein structures is often a challenge. If you’ve struggled with building smooth transition pathways for tasks like conformational analysis or free-energy simulations, you’re not…
Streamlining Molecular Animations with the Hide Effect
Bring Molecular Models to Life with the Pulse Animation
Preparing Protein Models for Conformational Pathway Searches.
Understanding File Attributes in SAMSON’s Node Specification Language (NSL)
For molecular modelers working on integrative projects, organizing and precisely targeting specific data within molecular models is a critical task. SAMSON, the cutting-edge platform for molecular design, leverages the Node Specification Language (NSL) to help users effectively control their datasets.…