Mastering Folder Attributes for Efficient Molecular Design
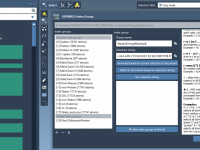
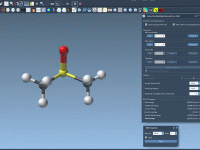
For molecular modelers working with complex structures and data, managing folders effectively can significantly streamline the design process. SAMSON, the integrative molecular design platform, provides a robust and intuitive system for working with folder attributes through its Node Specification Language…