Streamlining Molecular Disassembly: A Guide to the Disassemble Animation in SAMSON
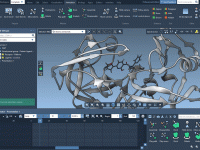
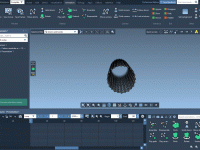
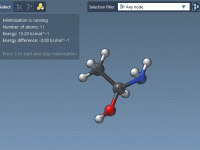
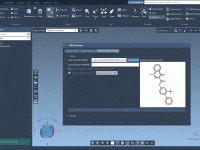
If you're a molecular modeler, you've likely encountered challenges when trying to visualize complex molecular systems. Whether it's understanding spatial arrangements or presenting your findings clearly, generating animations that simplify structural representations can be crucial in making your work accessible…