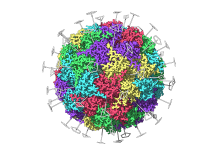
Reducing Docking Time with Smarter Search Domain Setup in Hex
A Simpler Way to Run AlphaFold-2 with SAMSON
From Click to Tube: Building Carbon Nanotubes with Your Mouse

Creating realistic models of nanostructures like carbon nanotubes (CNTs) can be time-consuming and technically complex. For molecular modelers and materials scientists, the ability to rapidly generate customizable CNTs is often crucial for prototyping device components, performing simulations, or illustrating theoretical…
Fade Away: A Simple Technique to Transition Molecular Visibility
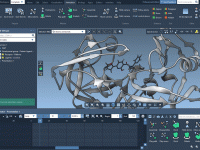
Struggling with presenting complex molecular scenes in a clean, digestible way? If you often build animations to showcase conformational changes, protein-ligand interactions, or intricate visualizations involving multiple overlapping layers, you’ve probably asked yourself: How do I control what’s visible and…
From Invisible to Visible: Using the ‘Appear’ Animation in Molecular Models
Tired of clicking atoms? Visualize biomolecular sequences interactively instead.
Managing Your Molecular Toolkit: Adding and Removing SAMSON Extensions Made Simple

When working on complex molecular modeling projects, researchers often need specific tools tailored to their workflows. One common challenge? Quickly finding and managing the right computational modules without interrupting your progress. That’s why flexible software like SAMSON—with customizable add-ons called…
Building Smooth Molecular Animations with Keyframes in SAMSON
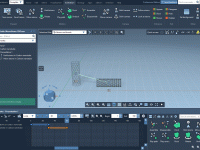
Creating scientific animations of molecular systems can be time-consuming, and even experienced modelers sometimes face challenges with accurately communicating dynamic behaviors. One common pain point many encounter is smoothly moving atoms or molecular fragments across frames without introducing artifacts or…