When (and Why) You Should Minimize Ligands Before Docking
Simplifying PMF Analysis with GROMACS Wizard: A Visual Workflow
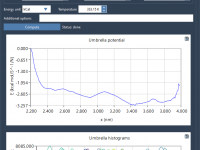
For researchers in molecular simulation, computing Potential of Mean Force (PMF) profiles can be an involved task—especially when dealing with data from multiple umbrella sampling simulations. Manual WHAM (Weighted Histogram Analysis Method) workflows in GROMACS often require scripting, data formatting,…
Making Your Molecular Models Speak: Customize Labels in SAMSON
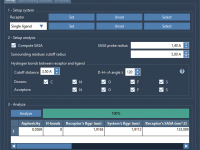
A Fast Way to Analyze Protein–Ligand Interactions in 3D Models
Quickly Locate Hidden or Visible Molecular Presentations in SAMSON
Keep It Out of Sight: How to Temporarily Hide Molecular Elements in Your Animations
In molecular modeling, attention to detail is everything. Whether you’re preparing a presentation, illustrating a mechanism, or crafting an educational video, highlighting specific structures—while letting others momentarily disappear—can improve clarity and focus. However, manually fading parts of a structure in…
Setting Up SAMSON for Faster Geometry Optimization with FIRE
Avoiding Common Pitfalls When Setting Up NVT Equilibration in Molecular Simulations
Running GROMACS Simulations in the Cloud Without Leaving Your GUI
For many molecular modelers, managing simulation workflows can become complex when switching between tools, environments, and machines. If you’re using GROMACS for molecular dynamics, you probably know the challenges involved with setting up simulations locally: installation and compilation, environment configuration,…