Building Lipid Bilayers Around Proteins with Molecular Box Builder
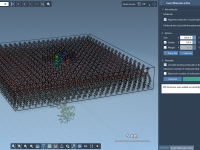
Modeling membrane proteins realistically often hinges on embedding them within a lipid bilayer. But placing lipids manually around a protein system can be time-consuming, potentially error-prone, and quite repetitive for simulations involving different lipid types or membrane compositions. Fortunately, Molecular…
Making Molecular Structures Easier to Understand with Visual Models in SAMSON
Making Light Nodes Work for You: A Guide to Visibility and Selection in SAMSON
A Simple Way to Visualize Protein Conformational Transitions in Seconds
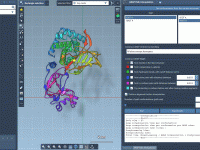
When exploring large-scale conformational changes in proteins, molecular modelers often face the same bottlenecks: visualizing smooth transition paths is time-consuming, and most available tools either require heavy computation or offer limited interactivity. This can slow down studies related to conformational…
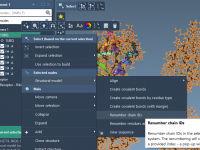
Avoid Topology Errors in Molecular Dynamics: Renumbering Chains and Residues for Martinize2
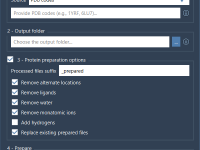
Cleaning Up Dozens of Protein Structures in One Go
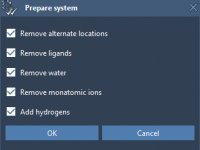
Avoid Common Pitfalls in Protein Docking: Setting Up Your System in SAMSON
Quickly Filter Your Molecular Render Presets in SAMSON with These NSL Attributes
Rendering molecular systems often involves combining clarity and aesthetic precision. In SAMSON, a powerful molecular design platform, users can assign different render presets to define how components are visually represented—whether it’s ribbons, surfaces, or atom spheres. But sorting or filtering…
Easily Animate Molecular Transformations Using Keyframes in SAMSON
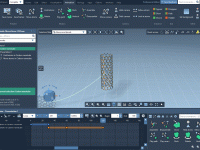
Creating animations of molecular behavior can be essential for illustrating dynamics, conformational changes, or transformations in chemical systems. However, many molecular modelers find this process time-consuming or difficult to control precisely. If you’re looking for a straightforward way to animate…