Author: OneAngstrom
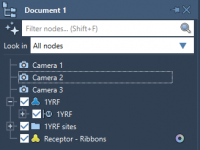
Effortlessly Fade Away Nodes in Molecular Designs with the Disappear Animation
Simplifying Python Package Management in SAMSON: An Overview
Streamlining Molecular Simulations: A Guide to Choosing Production Parameters in GROMACS Wizard.
For molecular modelers diving into production Molecular Dynamics (MD) simulations, choosing the right parameters can be daunting. Whether you’re simulating proteins, drug molecules, or other systems, getting these configuration details right ensures reliable results. In this blog post, we’ll unpack…