Author: OneAngstrom
Quickly Select Molecular Segments by Atom Counts, Charges, and More
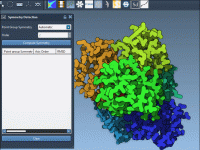
Which Symmetry Group Should You Trust?
Many molecular modelers working with large biological assemblies—such as viral capsids or homo-oligomeric protein complexes—find themselves asking the same question: What is the correct symmetry group governing my system? This is a common challenge, especially when automatic symmetry detection tools…
Need to Export Molecular Models Easily? Here’s How SAMSON Helps
Molecular modelers and computational chemists often face a common hurdle: exporting structures and visualizations in the right format for collaborators, publications, or simulations. With diverse tools requiring specific file formats like PDB, XYZ, CIF, or proprietary types, manually converting and…
When Symmetry Auto-Detection Isn’t Enough: How to Select Your Own Group in SAMSON
A Practical Way to Improve Ligand Unbinding Simulations with P-NEB
Keeping Views Consistent in Molecular Animations: How to Use ‘Hold Camera’ in SAMSON
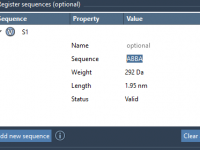
Design Smarter: How to Create and Use Monomer Sequences in SAMSON
Why the Shape of Your Simulation Box Matters More Than You Think
Easier Molecular Path Replay—In Reverse
For molecular modelers, visualizing conformational changes, docking simulations, or reaction mechanisms using trajectories is part of the daily routine. But what if you need to replay a molecular pathway—accurately—in reverse? Whether you’re trying to demonstrate molecular unbinding, study pathway reversibility,…