Author: OneAngstrom
What if your molecule looked back at you?
Quickly Find Charged Structural Groups in SAMSON Using NSL
Quickly Visualize Protein-Ligand Interactions After Docking in SAMSON
Ways to Select Paths by Atom Count in SAMSON
When and Why to Use Your Own GROMACS Version in SAMSON
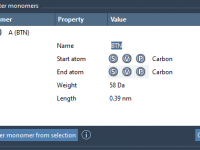
Managing Custom Monomers in SAMSON’s Polymer Builder: A Practical Guide
Adding and Managing Extensions in SAMSON Without the Guesswork
Fine-Tuning Molecular Movements: A Closer Look at Keyframed Atom Animation
For molecular modelers creating animations of dynamic systems, crafting smooth transitions between molecular conformations can be both a creative challenge and a technical hurdle. Whether you’re building an educational presentation or illustrating molecular events for a publication, precisely positioning atoms…