Mastering Movement in Molecular Models: A Guide to Move Editors
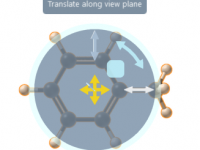
When designing or analyzing molecular structures, precise positioning and alignment are critical tasks that often demand intuitive tools. Whether you’re tweaking atomic placements, aligning meshes, or making large-scale adjustments, efficient movement tools can save you hours of frustration and refine…