Keep your view steady with the Hold Camera animation.
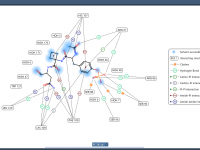
Mastering Protein-Ligand Interactions in 2D with SAMSON
Exploring Reverse Path Animations for Molecular Trajectories
One of the challenges in molecular modeling lies in effectively visualizing and analyzing the complex trajectories generated during simulations. Whether you’re cycling through conformations or revisiting molecular dynamics paths, the ability to reverse those pathways can provide useful insights. SAMSON's…
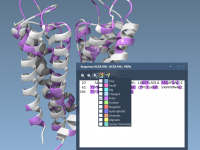
Mastering Residue Selection Across Views in SAMSON
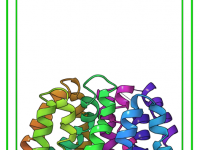
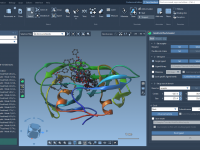
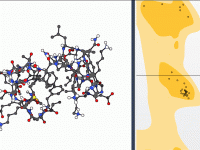
Enhance Depth Perception with Ambient Occlusion in Molecular Visualizations

Molecular modelers often face challenges when visualizing complex molecular structures. One frequent issue is accurately perceiving depth in intricate models, which can be critical for interpreting configurations and spatial relationships. If you’ve ever struggled with distinguishing overlapping regions in a…