Effortlessly Identify Hydrogen Bond Donors and Acceptors in Your Molecular Models
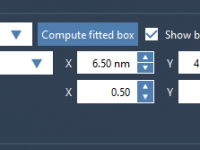
When modeling molecular systems, identifying hydrogen bond donors and acceptors quickly and accurately is often crucial. Whether you’re preparing structures for docking, analyzing interactions in a protein-ligand complex, or filtering atoms for visualization, being able to specify which atoms participate…