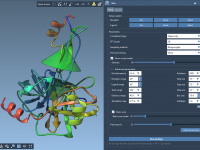
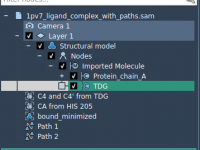
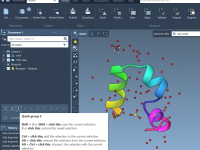
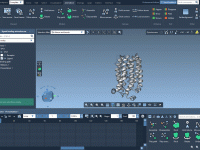
A straightforward way to export ligand trajectories from SAMSON
Why Understanding Node Types in SAMSON Can Save You Hours
Filtering Structures by Atom Count in SAMSON’s NSL
Switching Selections Fast When Modeling Molecules
From Conformations to Clearer Pathways: A Quick Optimization Strategy
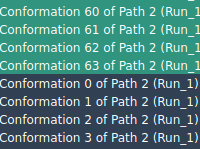
When modeling molecular systems, capturing the transition between two known structures—say, a ligand binding or unbinding event—often involves generating intermediate conformations. Although these conformations may be linearly interpolated or derived using dedicated algorithms like the Ligand Path Finder, they rarely…
Smooth Molecular Trajectories Without Scripting: A Close Look at the ‘Play path’ Animation
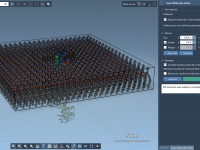
Building Lipid Layers Around Proteins Without the Headache
From SMILES to 3D Structures in Seconds: A Practical Guide for Molecular Modelers
A clearer way to filter node groups with NSL in SAMSON
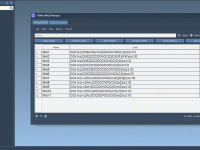
In structural modeling projects, distinguishing specific parts of complex systems can often be labor-intensive. Molecular modelers frequently need to filter or classify different parts of their system—such as ligands, water molecules, or fragments—based on attributes or naming conventions. SAMSON’s Node…